Post Electrophoretic Analysis Articles
S1 Mapping
Nuclease S1 will digest only ssDNA or ssRNA. If a duplex of DNA and/or RNA strands have single-stranded overhangs or unhybridized internal loops, these will be digested away. The remaining intact nucleic acid fragments represent regions of identity between two strands of the duplex. If one of the strands is labeled at one end, the length of the labeled fragment remaining after hybridization and nuclease digestion reflects the point on the probe where the two sequences diverge. This is the basis for S-1 mapping of transcriptional start sites (See figure below left). A probe is chosen that is complementary to the RNA and extends past the anticipated start site. The 5' end is 32P labeled, and chosen to fall within the coding region of the mRNA so that it will be protected from digestion. After hybridization, the 3' overhang of the probe is digested away, and the size of the remainder of the probe is accurately determined on a denaturing PAGE Gel. The distance between the known labeling site and the new end of the probe gives the transcriptional start site within 1 base.
S1 mapping can also be used to find intron sites (See figure below right). In this case, the probe is derived from genomic DNA and again labeled so that the labeled 3' end falls within a coding portion of the gene. Any intron in this construct will not find a homologous region in the RNA and will be cleaved by the S-1 nuclease. In this case, the size of the labeled remainder reflects the distance from the label site to the splice site.
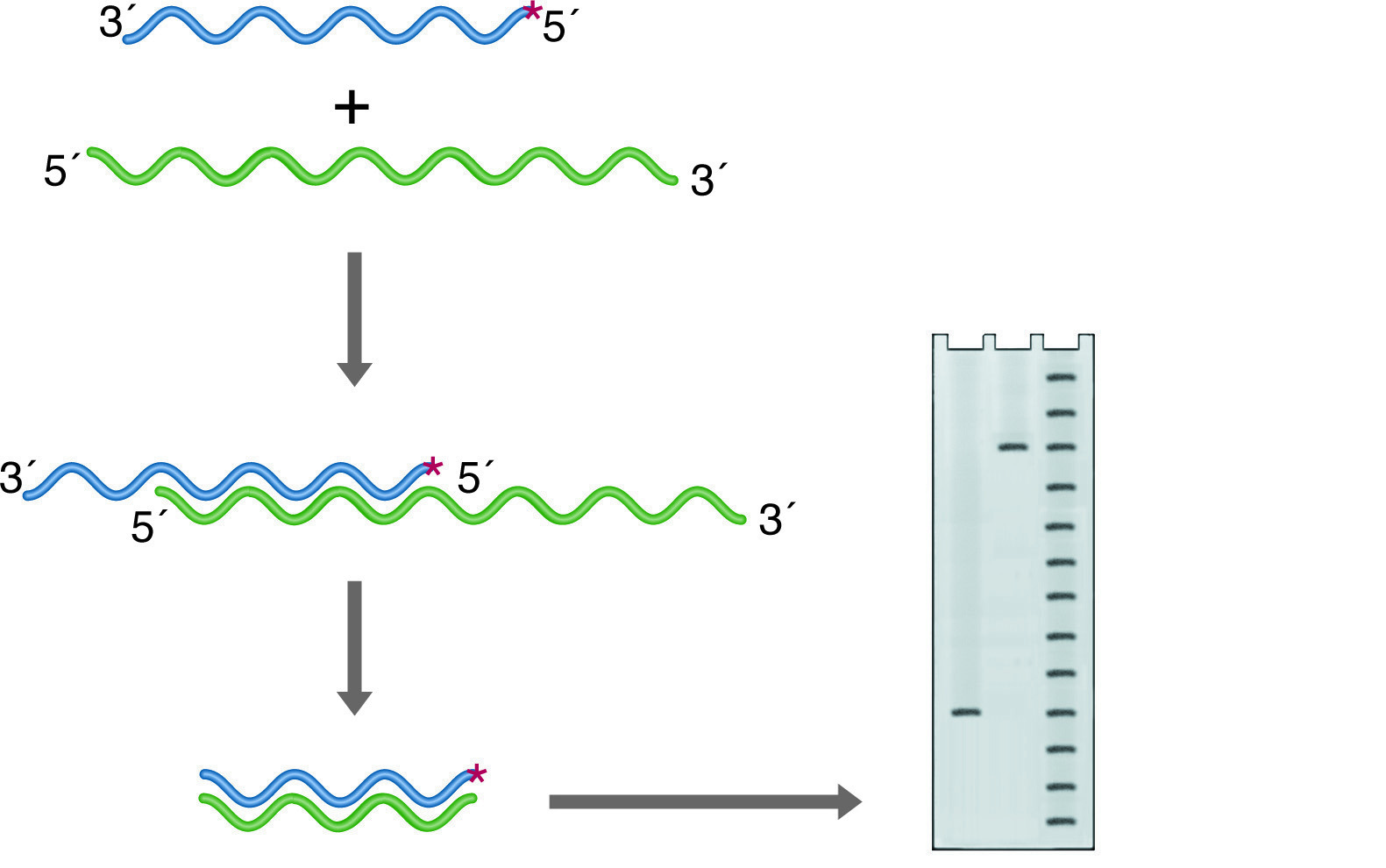
S-1 mapping of an intron site. The nuclease digests the unhybridized intronic DNA, which is not represented in the RNA. The size of the labeled fragment remaining indicates the distance between the 5' end of the probe and the intron position. Lanes: 1) Experimental, 2) Probe control, 3) Sequence ladder.
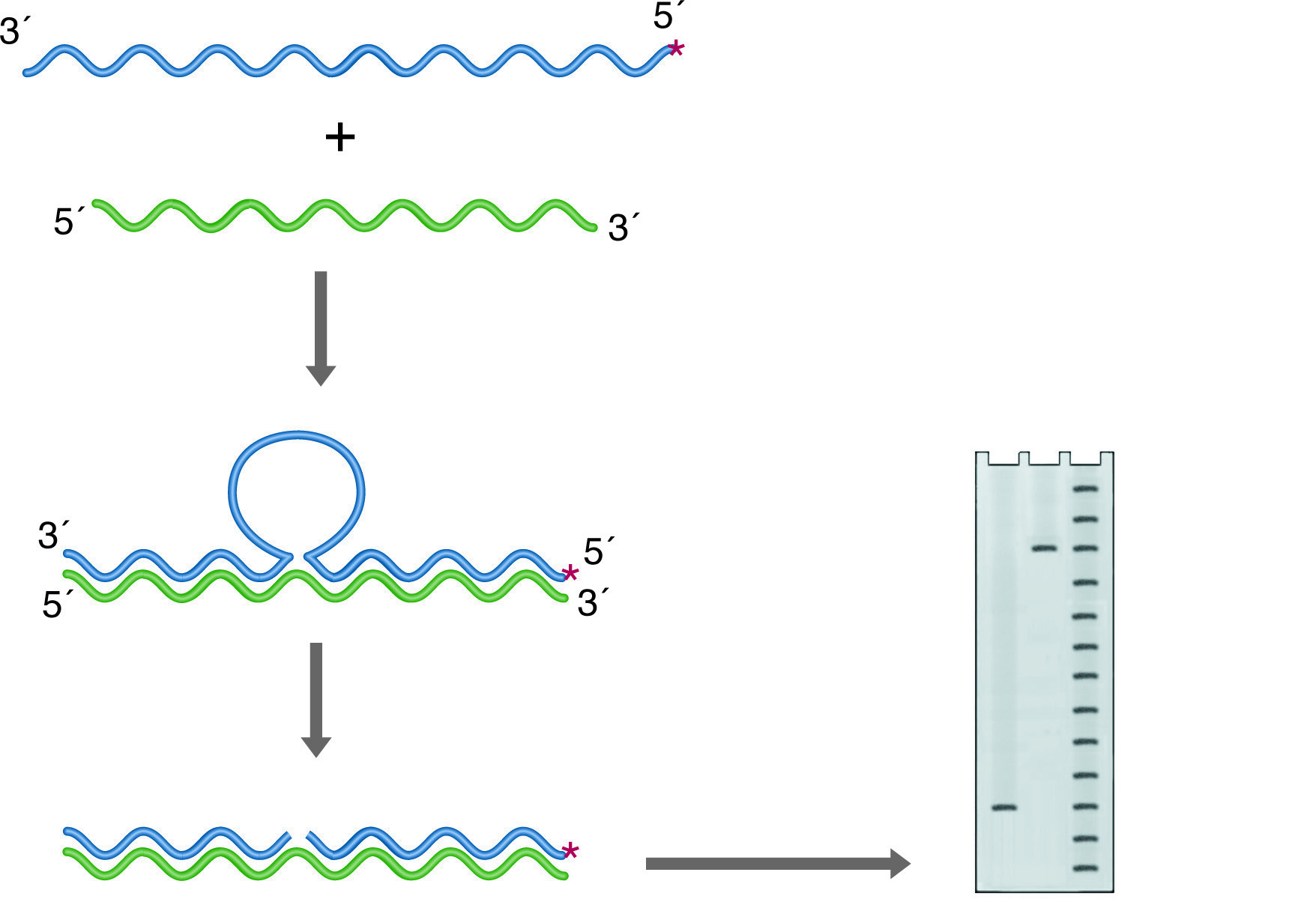
S-1 mapping of an intron site. The nuclease digests the unhybridized intronic DNA, which is not represented in the RNA. The size of the labeled fragment remaining indicates the distance between the 5' end of the probe and the intron position. Lanes: 1) Experimental, 2) Probe control, 3) Sequence ladder.
S-1 Mapping
NB: ALL REAGENTS AND GLASSWARE MUST BE RNase FREE OR DEPC TREATED.
- Probe preparation:
S-1 mapping requires a probe that is labeled at only one end. This can be accomplished in a number of ways, depending upon the available starting materials. Asymmetric restriction digestion of a cloned substrate can be manipulated to produce only one recessed 3' end, which can be extended by a Klenow fragment in the presence of labeled dNTPs. Alternatively, an end-labeled oligonucleotide may be extended with the Klenow fragment, and the duplex cleaved with a restriction enzyme to produce the required probe. However the probe is produced, it is generally purified on an alkaline agarose gel. Use a 1.2% low-melting gel. Locate the labeled band by autoradiography, excise, and trim it with the aid of a Geiger counter to remove excess agarose. Melt the excised band at 65°C, and add an equal volume of TE buffer. Add 10 µg of tRNA, and extract with 2x1volume of phenol (Tris equilibrated to pH 8). Ethanol precipitate the supernatant with 0.1 vol of 3M sodium acetate and 2 volumes of ethanol. Redissolve the pellet in 100ml of 0.3M sodium acetate. - Hybridization:
Add 1x105 counts of a labeled probe to 25-20 µg of RNA at 4°C. Add sufficient 0.3M sodium acetate to bring the volume to 100 µl. Add 300 µl of ethanol, pellet, and wash the precipitated DNA with 70% ethanol. Allow the pellet to air dry. Redissolve the pellet in 20 µl of S-1 buffer: 80% formamide, 40mM PIPES pH6.5, 400mM NaCl, 1mM EDTA. Heat to 65°C for 10 minutes and let stand overnight at room temperature. - S-1 Nuclease Digestion:
Add 300 µl of: 0.3M NaCl, 5mM ZnSO4, 50mM sodium acetate, pH 4.5; containing 300-500 Units of S-1 nuclease and 6 µg calf thymus DNA (denatured). Incubate at 30°C for 30-60 minutes. Stop the reaction with 100 µl of: 4M ammonium acetate, 25mM EDTA, 40 µg/ml tRNA. Add 1 ml ethanol, precipitate the DNA and redissolve in 5 µl of TE. Analyze 4 µl on a denaturing PAGE gel.
NEXT TOPIC: Ribonuclease Protection
