Post Electrophoretic Analysis Articles
Restriction Digest Mapping
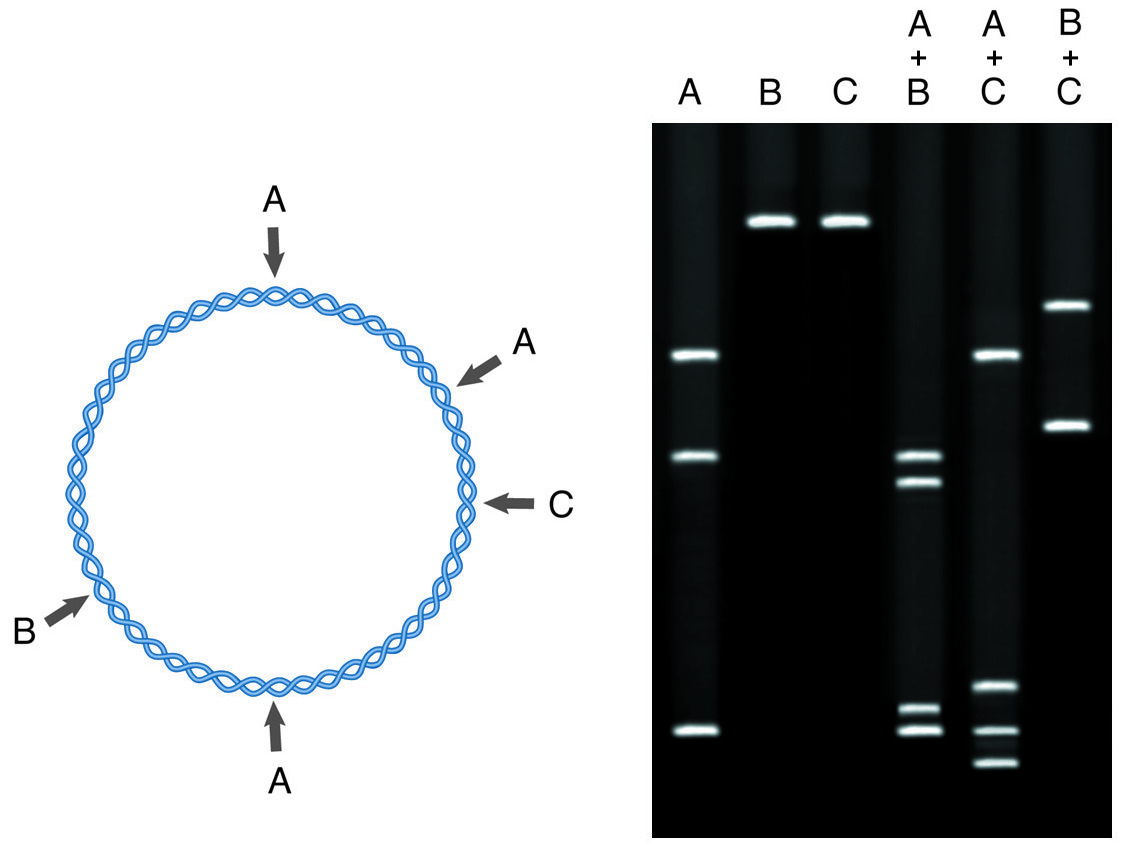
Restriction mapping involves the treatment of a DNA fragment with restriction enzymes both singly and in combination. The electrophoresis of cleavage products yields a map of the DNA in terms of restriction sites.
Restriction endonucleases are enzymes that cleave double-stranded DNA at specific sites, generally 4, 6 or 8 base palindromic sequences. Because, in action, the enzymes are sequence-specific, each piece of DNA has a recognizable pattern (or map) of restriction sites. This map serves as an easily identifiable “fingerprint” whereby the identity of a piece of DNA can be established without recourse to sequencing or blot hybridization. A map of restriction sites is also essential for planning subcloning experiments, in which a large piece of DNA is cut into smaller fragments for more convenient analysis.
To perform a restriction mapping experiment, 2-10µg of sample DNA is digested to completion in a set of separate reactions with 5-10 different restriction enzymes. These reactions provide the primary map information, giving the distances between the restriction sites and the ends of the DNA molecule, or revealing the existence of multiple sites for one enzyme. This information is extracted by running the digestion reactions on an agarose gel vs standards of known size, to determine the size of each restriction fragment generated.
In the second stage of restriction mapping, the DNA is digested with pairs of enzymes (double digests) selected from the enzymes used in the single digests. The sizes of the fragments generated indicate the relative positions of the restriction sites for the two enzymes involved. Generally, the second round of double digests is then carried out to resolve the remaining ambiguities in the map.
NEXT TOPIC: DNA/RNA Purification from Agarose Gels
