Post Electrophoretic Analysis Articles
Automated Sequencers
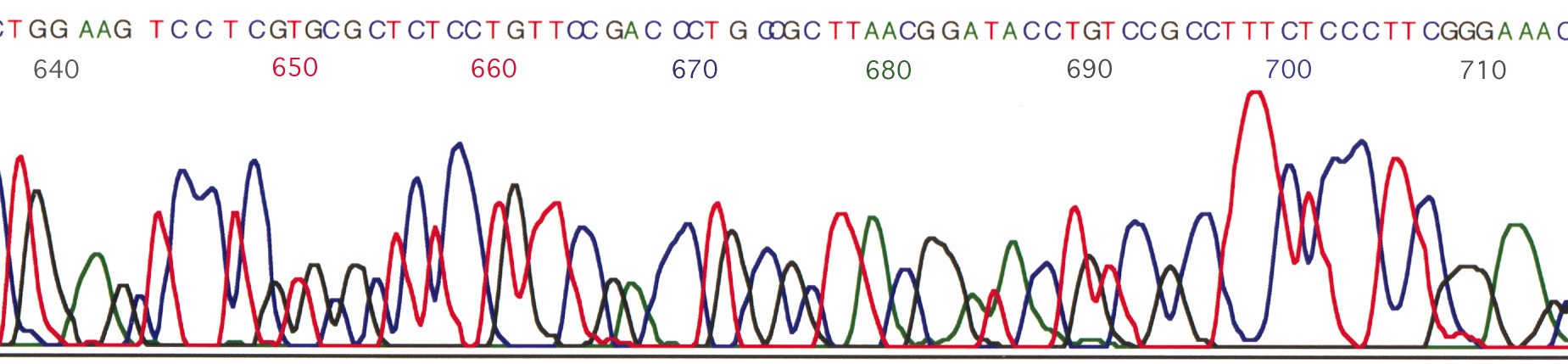
Automated sequencing systems make use of fluorescent dye labeling, in combination with laser scanning and computerized data acquisition and processing to carry out the electrophoresis of up to 96 sequencing reactions on a single gel, and read over 1,000 bases from each reaction. A single run on an automated sequencer can thus produce as much data as 40 manual gels.
Reactions for automated sequencing use the same Sanger dideoxy chemistry as manual sequencing systems. The key difference lies in how the fragments are labeled for later detection. Instead of radiolabeling, highly fluorescent dye molecules are linked either to the primer (dye-primer) or the dideoxy NTP's (dye terminator). Excess dye is removed in a post-reaction cleanup step, and the products are separated on a denaturing polyacrylamide gel as used for manual sequencing. The gel is run while mounted on a detector, which constantly scans a laser across all lanes at the bottom of the gel. When a labeled band crosses this detection zone, its fluorescence is detected and recorded. The pattern of fluorescent flashes over time is interpreted into a DNA sequence using a computer.
One of the strengths of this system is that multiple colors of fluorescent dye are available (note that many of these dyes are fluorescent in the UV or IR regions, so the term "color" is used as a shorthand for "fluorescent at a different wavelength." In general, a different color is used for each ddNTP reaction. As a result, all 4 reactions can be run on a single lane. Each band is identified with a position in the DNA by its elution time, and with a base by its color.
Although automated sequencing uses unique labeling and processing techniques, the electrophoretic separation at its core is essentially the same as that for manual sequencing. The key differences are in the much higher standard of purity required, and the focus on larger DNA fragments. Automated runs may take as long as 12 hours to complete. The gel must remain intact and unaltered during that time, at elevated temperatures. Additionally, the use of laser scanning means that ultra low levels of contaminant will show up as elevated background fluorescence. Also, gel results are processed according to parameters set in the computer software, which must be reproducible from gel to gel. All of this demands that the acrylamide, urea and buffers used be of exceptionally high purity.
For most automated sequencers, the best matrices are modifications of the traditional 19:1 manual sequencing gels. The need for modification arises because, while manual sequencing generally aims to read 400 bases at most from a reaction, automated sequencing in some machines allows over 1000 bases to be read. In these machines, a more open pore structure is required to allow the resolution of the larger fragments. For most purposes, National Diagnostics recommends the use of SequaGel-UreaGel 6 in the ABI-373 or SequaGel XR in the ABI-377, and SequaGel XR in the LICOR sequencers.
NEXT TOPIC: Differential Display
