Gel Electrophoresis of Proteins
Measuring Molecular Weight with SDS-PAGE
The mobility (Rf) of a molecule in gel electrophoresis is determined by its free solution mobility, Y0 (= mobility in a gel of zero %) and the sieving action of the gel matrix. In denaturing protein electrophoresis, the addition of SDS to the electrophoresis buffer uniformly coats the proteins with negative charges, equalizing the charge to mass ratio for all proteins, thus making Y0 he same for all species. In this case, relative mobilities are determined solely by the sieving action of the gel. This sieving action is proportional to the molecular weight (MW) of the particular protein. Theoretical treatments suggest that the logarithm of Rf should vary with MW, but most users use an empirical plot of the logarithm of MW vs Rf for several standards of known molecular weights to determine the molecular weights of unknowns.
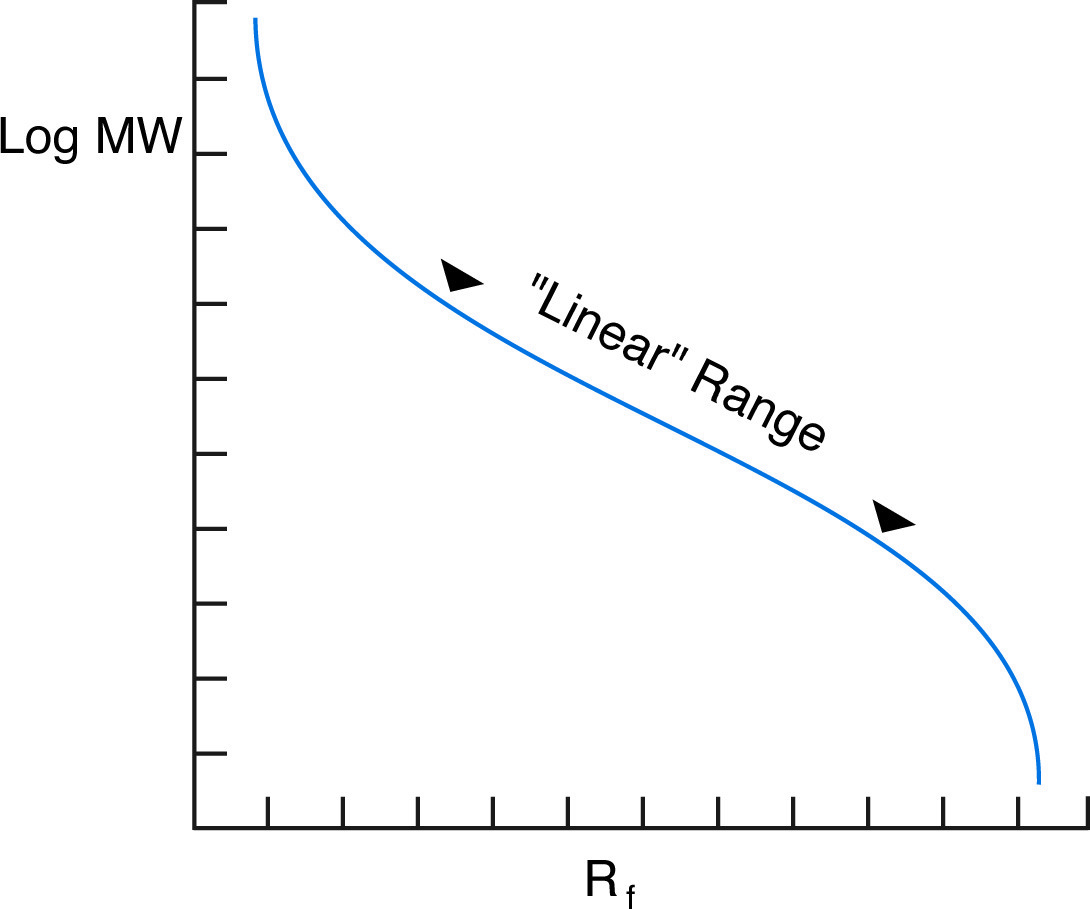
Although the overall graph of log MW vs. Rf is sigmoidal, it is nearly linear for a range of molecular weights depending on the percentage monomer of the gel.
In practice, the proportionality of log(MW) vs Rf holds true for most proteins, provided they are fully denatured and provided the gel percentage has been chosen to match the molecular weight range of the sample. In fact, the actual plot of log(MW) vs Rfis sigmoidal (see figure below), because at high MW, the sieving effect of the matrix is so large that molecules are unable to penetrate the gel, while at low MW, the sieving effect is negligible, and proteins migrate almost at their free mobility, which in SDS is independent of MW.
Given an appropriate selection of gel % (see table below) and a protein that displays near-ideal behavior, molecular weights can be determined to be within 5 - 10%. Molecular weights of non-ideal proteins can be determined by the use of Ferguson Plots, a technique that employs native protein electrophoresis.
Determining Molecular Weight with Gradient SDS Gels
If a gradient of acrylamide concentration is introduced into SDS PAGE, larger ranges of proteins may be analyzed on the same gels, with greater resolution. The complexity of the relationship between migration and molecular weight is dependent upon the shape of the gradient. The overall equation is of the form log(MW) α log(P), where P is the concentration of acrylamide at the band position. A graph of log(MW) vs log(P) is linear and allows the determination of MWs from a set of standard protein positions. For linear gradient gels, the percentage of acrylamide is proportional to the position in the gel, so log(MW) will be proportional to log(band position). Therefore, a graph of log(MW) vs log(Rf) for a set of standards will be linear, and Rf values for unknowns can then be converted to MW values. On a 3 - 30% gradient gel, a range of proteins that differ in MW by up to 100 fold can be resolved and MW's determined.
NEXT TOPIC: Peptide Mapping
- Sample Preparation for SDS-PAGE
- Sample Preparation for Native Protein Electrophoresis
- Peptide Mapping
- Native Protein Electrophoresis
- Measuring Molecular Weight with SDS-PAGE
- Isotachophoresis
- Isoelectric Focusing
- Immuno-Electrophoresis / Immuno-Diffusion
- Gel Preparation for Native Protein Electrophoresis
- Gel Electrophoresis of RNA & Post Electrophoretic Analysis
- Denaturing Protein Electrophoresis: SDS-PAGE
- Casting Gradient Gels
- Activity Stains
