Post Electrophoretic Analysis Articles
SSCP Analysis
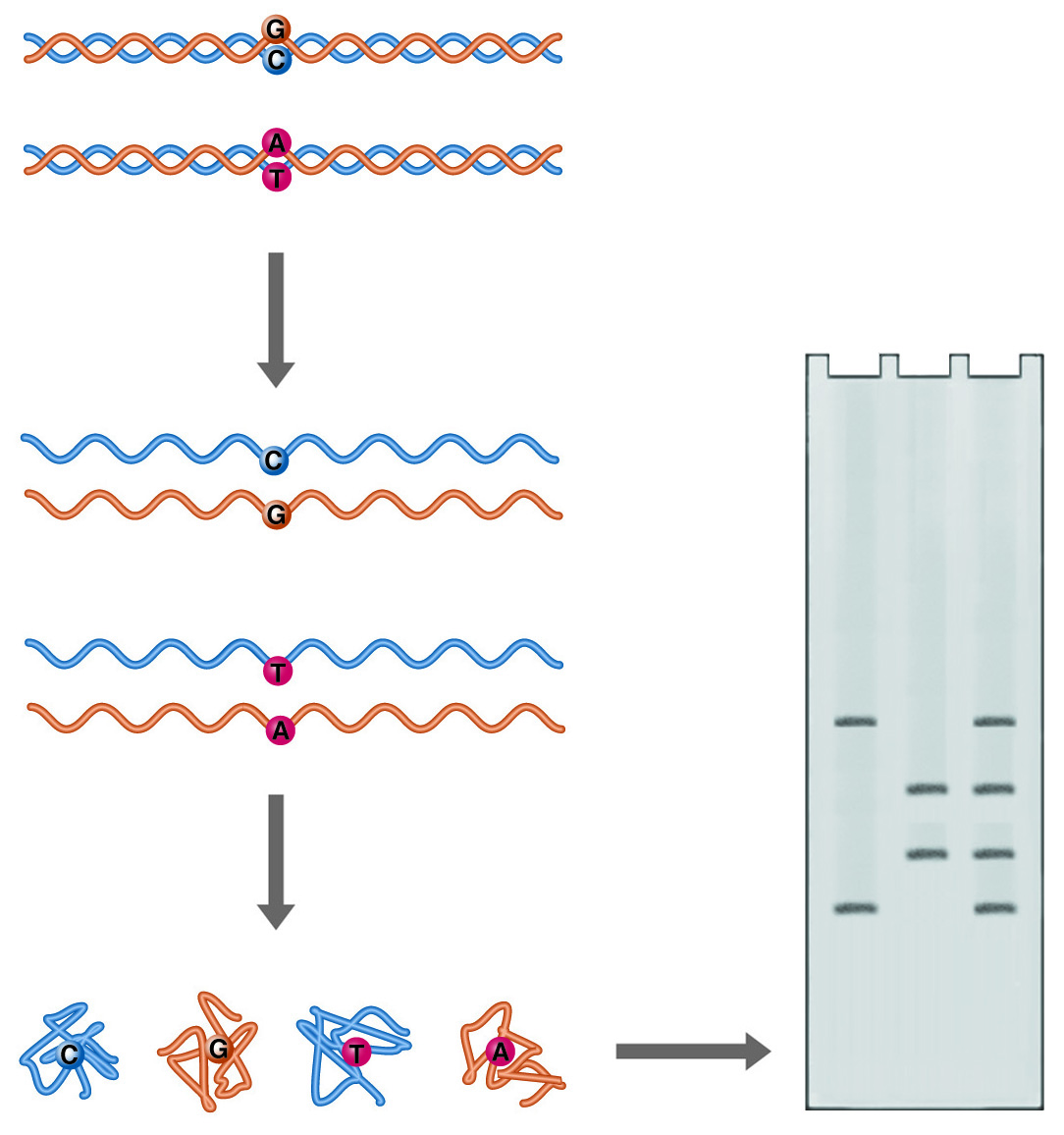
SSCP (Single Strand Conformational Polymorphism) Analysis. Single point mutations can cause major differences in the folded form of single-stranded DNA. These differences can be detected as differences in electrophoretic mobility.
Single-stranded DNA can adopt multiple conformations under non-denaturing conditions. In the absence of a complementary strand, DNA will anneal short internal complementary sequences, forming a complex "knot." A DNA molecule follows a complex path of folding to reach its final form, influenced by the solution environment and temperature. The path consists of a series of annealing steps, each stabilizing and to some extent directing the next. Minor alterations in the sequence of the DNA will disrupt the annealing process and result in a different final shape. The compactness of these structures will determine how fast the single-stranded DNA migrates through a non-denaturing gel. Such differences in electrophoretic mobility between nearly identical strands serve as the basis for the technique known as single-strand conformation polymorphism (SSCP) analysis, and like heteroduplex analysis, it is a rapid method for mutational screening.
Samples are denatured with heat and then rapidly cooled. Rapid cooling favors self-annealing because insufficient time is allowed for complementary strands to collide and orient for duplex formation. The renatured samples are analyzed on a gel opposite the control DNA. As with heteroduplex analysis, all fragments must be the same length. Mutant samples will show mobility different from the control DNA. The gel matrix used must be optimized for the resolution of DNA conformers of the same length. Various combinations of acrylamide and bis-acrylamide are mentioned in the literature, at ratios from 29 to 1 to 50 to 1, and at percentages from 4 to 8. National Diagnostics' SequaGel MD is optimized to provide superior results in both heteroduplex and SSCP analysis.
SSCP Analysis Procedure
GEL PREPARATION
(for 0.4 mm thick gel):
- Combine 25 mL SequaGel MD and 6 mL 10X TBE in an Erlenmeyer flask. Fill to 100 mL with deionized water. Mix thoroughly. Prepare 0.6 X running buffer by diluting 60 ml of 10X TBE stock to 1 L with deionized water.
- Cast the gel by adding 40 µl TEMED and 400µl of freshly prepared 10% ammonium persulfate to the solution. Swirl gently to mix. Using standard acrylamide procedures, pour the gel solution into the cassette, insert the comb (inverted if using a sharks tooth comb), and allow polymerization at room temperature for a minimum of 60 minutes. Attach the gel cassette to the electrophoresis apparatus.
SAMPLE PREPARATION
- Attempt PCR amplification. Note: PCR conditions should be optimized for desired PCR product before SSCP analysis is undertaken. It is recommended that the minimum number of PCR cycles be used on a purified, salt-free template, and that reagent and primer concentrations be optimized. If radiolabeling is going to be utilized instead of silver staining, end-labeled primers may be used, or an a-32P dNTP may be included in the PCR amplification.
- After PCR thermal cycling, add EDTA to a final concentration of 5 mM (1µl of 0.5 M EDTA per 100 µl reaction) to inactivate the Taq DNA Polymerase.
ELECTROPHORESIS:
- Rinse the gel wells with running buffer. The shark's tooth comb should be reinserted so that it just touches the surface of the gel, and 1 to 3 µl of the sample should be loaded.
- Run the gel at a constant power of 6-8 watts for 14 hours.
- If the DNA was radiolabeled, transfer the gel to Whatman 3MM filter paper, place it on a flat surface, and cover it with plastic wrap. Dry the gel and, using standard technique, expose it to X-ray film. Silver staining may be used for detection. Ethidium bromide is not effective at detecting single-stranded DNA.
NEXT TOPIC: Agarose Gel Electrophoresis of DNA & RNA-An Introduction
